Welcome to AP-Nimbus’s documentation¶
| Project Home: | https://github.com/CardiacModelling/ap-nimbus |
|---|---|
| Documentation: | https://ap-nimbus.readthedocs.io/ |
| Created: | Aug 07, 2023 |
| Version: | 1.0.1 |
IMPORTANT¶
This activity represents the next step in the development of the original AP-Portal work – towards a container-based / cloud solution.
AP-Nimbus is available at https://github.com/CardiacModelling/ap-nimbus and is
being developed by the University of Nottingham’s
School of Mathematical Sciences .
Preamble¶
- Unlike AP-Portal, this work includes the installation of
ApPredict, the cardiac simulation software.- Also unlike
AP-Portal, this work, by nature of containerisation, means thatAP-Nimbuswork does not embody a single application, it is instead a collection of containers where each can operate in isolation, e.g. as a standaloneDockerorSingularitycontainer, or alternatively, orchestrated in a microservice architecture (e.g.KubernetesorDocker Compose).
Singularity¶
This documentation predominantly covers Docker container environments, however
it has been relatively straightforward to create Singularity
containers (e.g. singularity build app-manager.img docker://cardiacmodelling/ap-nimbus-app-manager:0.0.10)
and use those [1].
Sample invocation scripts can be found at ap-predict-online’s app-manager –> tools section.
Diagrammatic Representation¶
For the role each of the containers has in the overall AP-Nimbus activity please
see Activity Overview.
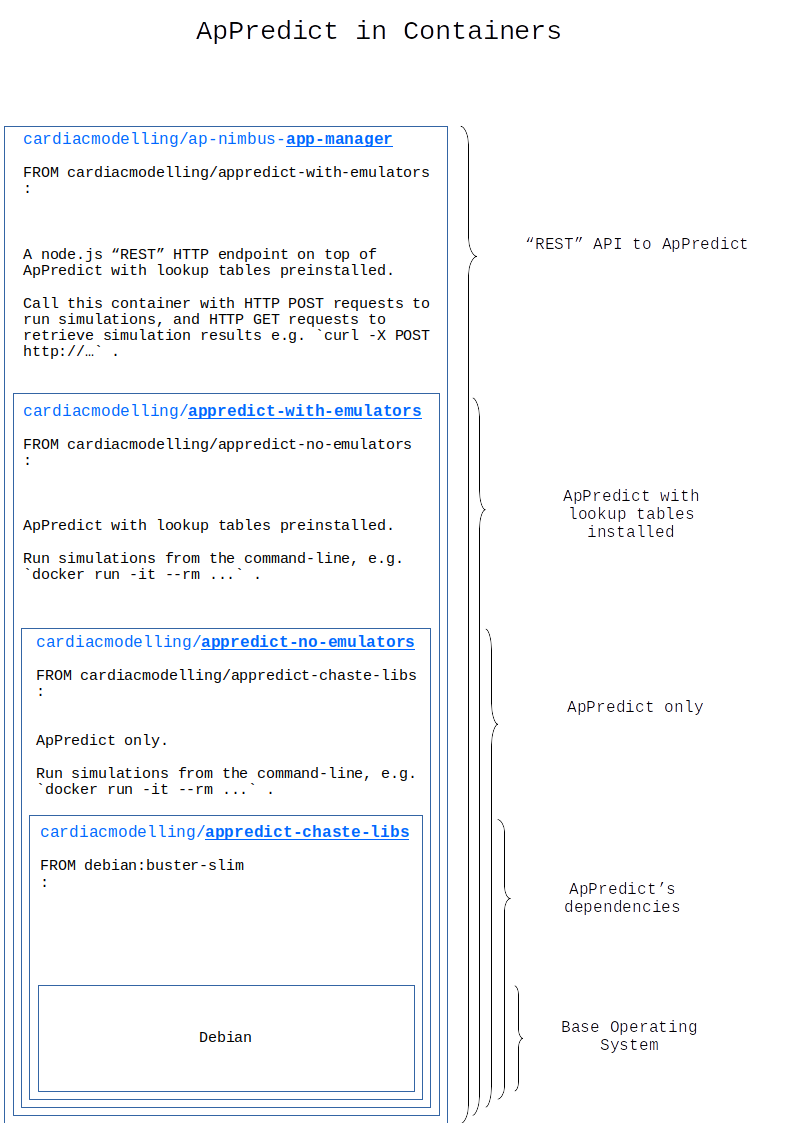
ApPredict containers¶
ApPredict is the underlying cardiac simulation engine.
Building or installing ApPredict is a complex and time-consuming
process and by distributing in container form it’s possible to have it installed
in a fraction of the time [2].
AP-Nimbus containers¶
The following illustrates a Docker container setup.

It is equally feasible to run as .. :
- Containerised
docker run ..a singlecardiacmodelling/ap-nimbus-app-managercontainer and call it with HTTPPOSTandGETrequests, or;docker run ..either of thecardiacmodelling/appredict-with-emulatorsorcardiacmodelling/appredict-no-emulatorscontainers directly from a CLI to run their internalApPredicts, or;- Non-containerised
- Running the various components without using containers is technically possible, but not reccomended or supported. See Developer Section for suggestions on how to develop the components using containers.
See also
For instructions on how to run containers, see the more detailed section on Running.